Projects
Sphingolipid Metabolism in Hepatotoxicity
Sphingolipids (SPs) consist of a highly diverse class of lipids that function both as structural components of cellular membranes and signaling molecules eliciting apoptosis, proliferation, gene expression, and immune response. This project aims to comprehensively characterize SP structure and abundance by integrating chromatography, adduct formation, ion mobility, and tandem mass spectrometry. This multidimensional approach provides in-depth structural information such as acyl chain configuration, double bond location, and distinction of isomeric species. Further, we are anchoring structure to accurate quantitation. We are currently applying this approach to investigate the role SP metabolism plays as a molecular marker for hepatotoxicity in a drug-induced liver injury model system.
References:
Li L, Wang H, Jones JW. Sphingolipid metabolism as a marker of hepatotoxicity in drug-induced liver injury. Prostaglandins Other Lipid Mediat. 2020 Sep 30;151:106484. doi: 10.1016/j.prostaglandins.2020.106484. PubMed PMID: 33007444.
Tran A, Wan L, Xu Z, Haro JM, Li B, Jones JW. Lithium Hydroxide Hydrolysis Combined with MALDI TOF Mass Spectrometry for Rapid Sphingolipid Detection. J Am Soc Mass Spectrom. 2020 Oct 30;. doi: 10.1021/jasms.0c00322. PubMed PMID: 33124427.
Ether Phospholipids in Neurodegenerative Diseases
This project focuses on elucidating ether phospholipid dysregulation in neurodegenerative diseases such as traumatic brain injury (TBI). Due to their unique structure, ether phospholipids, specifically plasmalogen glycerophosphoethanolamine (PE), play a significant role in membrane dynamics, cell-cell interactions, and cellular signaling. Our research interests aim to understand how changes in PE composition can provide mechanistic insight into injury progression. We incorporate multidimensional approaches such as liquid chromatography, ion mobility, and tandem mass spectrometry to comprehensively evaluate changes to PE structure and abundance. Furthermore, we are interested in understanding how such changes may alter important organellular functions during TBI pathogenesis.
References:
Mehrabani-Tabari A, et al. Peroxisomal ether-glycerophospholipid synthesis is dysregulated after TBI. J Lipid Res. 2025 May 7;:100821. doi: 10.1016/j.jlr.2025.100821.
Ji Y, et al. Development and evaluation of a liquid chromatography-tandem mass spectrometry method for simultaneous measurement of toxic aldehydes from brain tissue. J Chromatogr B Analyt Technol Biomed Life Sci. 2024 Jul 15;1242:124208. doi: 10.1016/j.jchromb.2024.124208.
Morel Y, Jones JW. Utilization of LC-MS/MS and Drift Tube Ion Mobility for Characterizing Intact Oxidized Arachidonate-Containing Glycerophosphatidylethanolamine. J Am Soc Mass Spectrom. 2023 Aug 2;34(8):1609-1620. doi: 10.1021/jasms.3c00083.
Morel Y, Hegdekar N, Sarkar C, Lipinski MM, Kane MA, and Jones JW. Structure-Specific, Accurate Quantitation of Plasmalogen Glycerophosphoethanolamine. Submitted and under review. June 2021.
Jones JW, Sarkar C, Lipinski MM, Kane MA. Detection and Structural Characterization of Ether Glycerophosphoethanolamine from Cortical Lysosomes Following Traumatic Brain Injury Using UPLC-HDMSE. Proteomics. 2019 Sep;19(18):e1800297. doi: 10.1002/pmic.201800297. PubMed PMID: 30790445.
Lipid Structure/Function in Bacterial and Viral Pathogenesis
Lipids are important biomolecules in both the host cell and the pathogen. From a pathogenic perspective, lipids serve a protective role to safeguard against external pressures (e.g., harsh temperature or pH levels, and host immune defenses), and pathogenic lipids also play an active role in host recognition and subsequent entry into the host cell. Conversely, host cell lipids are vital to proper cell function and are at the forefront to the host’s cell response to interacting with a pathogen. We are interested in investigating the role lipids play at the interface of pathogenesis. This work is done by characterizing membrane lipids from enveloped viruses for identification of biomarkers to detect infections and as drug development targets. We are developing discovery and quantitative mass spectrometry methods to accomplish this work.
Tran A, Monreal IA, Moskovets E, Aguilar HC, Jones JW. Rapid Detection of Viral Envelope Lipids Using Lithium Adducts and AP-MALDI High-Resolution Mass Spectrometry. J Am Soc Mass Spectrom. 2021 Apr 22;. doi: 10.1021/jasms.1c00058. PubMed PMID: 33886294.
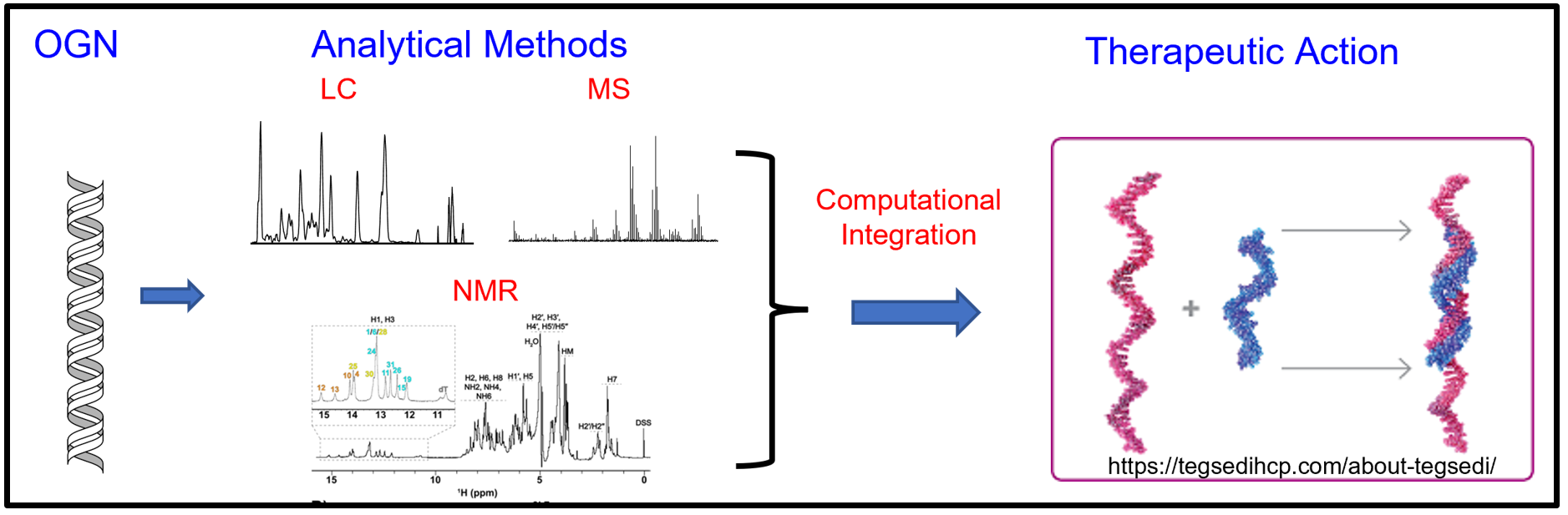
Combined Analytical and Computational Approach to Comprehensively Characterize Diastereomer Composition in Oligonucleotide Therapeutics
Short (synthetic) oligonucleotide (OGN) therapeutics are an emerging class of biopharmaceuticals to treat and prevent a wide variety of human diseases, including those deemed “undruggable” by traditional small molecule and protein-based approaches. This therapeutic platform is highly attractive due to the specificity for modulation of gene expression and improved PK, PD, and biodistribution properties achieved by chemical modification. In contrast to native nucleic acids, which are prepared enzymatically, short OGN therapeutics are prepared chemically via solid phase chemical synthesis. This process imparts chemical modifications to nucleotide bases, sugar moieties, and internucleotide linkages. Of the several different varieties of OGN therapeutics, antisense oligonucleotides (ASOs) and small interfering RNAs (siRNAs), are at the forefront in drug discovery and FDA drug approval. One chemical modification commonly employed for OGN therapeutics involves the conversion of the natural phosphodiester nucleotide linkage into a phosphorothioate (PS) linkage. This modification confers nuclease resistance and enhance protein binding. A by-product of the PS linkage is a newly introduced chiral center creating 2n diastereomers, where n is the number of PS linkages. A 20-mer ASO drug (e.g., TEGSEDI) potentially yields 219 (524,288) stereoisomers. Characterization of PS stereoisomer composition for OGN drugs is highly consequential due to the potential to alter pharmacological effects and uncharacterized and/or uncontrolled PS diastereomer compositions adversely impacting active ingredient bioequivalence. Despite the increasing popularity of OGN drugs, there is a pressing need to develop high-resolution analytical methods that can comprehensively characterize the diastereomer composition in OGNs for quality control purposes in manufacturing and generic drug development.
Becette OB, Tran A, Marino JP, Jones JW, Brinson RG. Rapid identification of short oligonucleotide impurities using lithium adduct consolidated MALDI-TOF mass spectrometry. Int. J. Mass Spectrom., 481, 2022, 116913. ISSN 1387-3806. Doi: 10.1016/j.ijms.2022.116913.
Becette OB, Tran A, Jones JW, Marino JP, Brinson RG. Structural Fingerprinting of siRNA Therapeutics by Solution NMR Spectroscopy. Nucleic Acid Ther. 2022 Aug;32(4):267-279. doi: 10.1089/nat.2021.0098.
We are developing a multidimensional analytical and computational approach to integrate high-resolution techniques (mass spectrometry and NMR) with machine learning. These approaches are being paired with synthetic chemistry and pharmacological assays to investigate how PS stereochemistry affects biological activity.
This project involves a collaborative effort across multiple disciplines: 1. Chromatography, Ion Mobility and Mass Spectrometry: Jones Lab (UMB SOP PSC) 2. NMR: Brinson Lab (NIST/IBBR) 3. Bioinformatics: Cummings Lab (UMD Biology) 4. Synthetic Chemistry: Fletcher Lab (UMB SOP PSC) 5. Pharmacology: Wang Lab (UMB SOP PSC)